This paper1 is mainly the work by the student Sebastian Machera, who spend some time working in the Surface Nanoengineering group. Together with Dr Katarzyna Szot-Karpińska, he was working on phage display technology to make bacteriophage express peptides that could recognise specific targets. In this case, the model target was Troponin T.
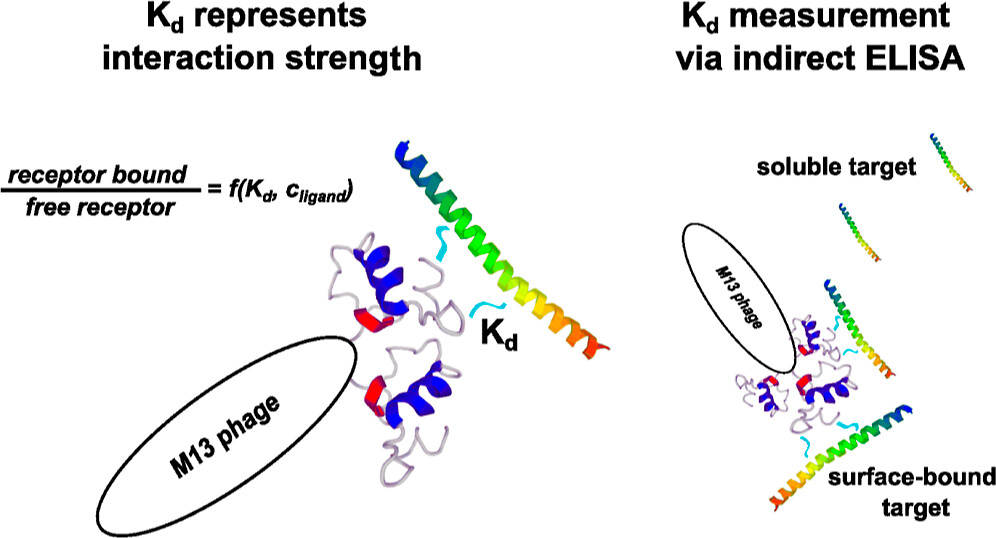
As the title implies, the goal here was to determine the dissociation constant for a binding event based on ELISA measurements. There are well-established procedures in the literature for how this can be done. However, mostly for monovalent receptors. Here, a bacteriophage displaying target specific peptides was the receptor, and this bacteriophage can display more than one receptor. Sebastian derived the expression for calculating the Kd value for polyvalent receptors and applied this to his specific bacteriophage. The issue here is that every bacteriophage (or other polyvalent receptor) can bind either one, two, or more target molecules, and getting from that varying binding to a single number for Kd require some care. Also, the way ELISA works here, means that any completely saturated phage will not be detected.
The article also contains a lot of experimental work on biopanning for bacteriophages with specific affinities and measuring this using ELISA. All in all, a massive amount of work has gone into the paper. Martin’s contribution was helping Sebastian find the numerical solutions to the equations he had developed and also helping with some editing to make the derivation understandable for someone who didn’t spend months getting familiar with the issue.
- S. J. Machera, J. Niedziołka-Jonsson, M. Jonsson-Niedziółka, K. Szot-Karpińska
Determination of the Dissociation Constant for Polyvalent Receptors using ELISA: a Case of M13 Phages Displaying Troponin T-specific Peptides, ACS Omega (2023) (link)